*** Work in progress ***
These programs use the custom MCS wrapper library for MySQL implemented in dynamically loadable modules (DLMs). It requires then that you have MCS configured with the option --enable-idl.
Two contributed libraries allow an even easier writing of IDL procedures and functions to interact with the MySQL server (mcs_usrlib.pro) and to produce and plot HEALPix sky maps (mcs_healplib.pro).
HEALPix team provided IDL routines are documented here. They are also available from the top Soft directory (here or the MCS web site here).
Install them in a directory listed in the IDL_PATH and all should be fine.
In order to be able to use the IDL wrapper routines you need to run (only once!) a MCS specific command from the directory where you intend to use them. This is required to create logical links with the DLM file and shared library needed by IDL at runtime.
For example from the Soft/IDL:
shell>
cd Soft/IDL
idl2mcs
Press the ENTER key
You’ll see that 2 links are created:
shell> ls -l idl2mcs*
... idl2mcs.dlm -> /usr/local/share/mcs/client/idl2mcs.dlm
... idl2mcs.so -> /usr/local/lib/idl2mcs.so
Can edit and have a look to the .dlm file (it’s ASCII text).
The available demo programs only show part of the capabilities of the libraries. If you are an IDL programmer it should be easy to edit the source code and get inspired to implement your own custom programs.
All the demo programs have a description header. Please read it. Also note that the mcs_healplib.pro invokes itself mcs_usrlib (@mcs_usrlib in the first line) so when you write procedures requiring HEALPix facilities, just add as first line @mcs_healplib. Briefly:
- myquery1.pro: Simple query using the MCS to IDL wrapper. mcs_usrlib.pro is NOT used.
- dbtest1.pro, dbtest2.pro: simple queries using the MCS to IDL wrapper via the mcs_usrlib.pro
- dbexample1.pro: very simple demo pro to perform queries via the mcs_usrlib functions and programs:
- perform a SELECT query on the INFORMATION_SCHEMA DB,
- create the mpe2018db.test_mcs table and perform various INSERT, UPDATE, REPLACE and SELECT queries,
- show how to get queries metadata info,
- show how to get the query output in various formats (structure or 2-d array).
Note: because we use DBs that do not require any particular user privilege, you can just hit RETURN when user/password are requested.
- simpleqry.pro: use the simpleBSC table in the mpe2018db DB to perform a SELECT query and get the result in various formats. Uses the mcs_usrlib routines: DBExec_Qry, DBExec_TabQry and DBExec_ShowQry, DBTabFLT.
- dbexample_reg.pro: use by default the HTM 6 regions table in the mpe2018db DB. If notinstalled, get it from regions.sql.gz. It makes use of the following mcs_usrlib routines: DBExec_Qry, TabStruct, TabFill, FieldInfo, DBFree. It accepts the following input keywords:
Host: The DB server host name (def. localhost). User: The user name to use for DB access. Password: The password to use for DB access. DBName: The database name to access. Query: The complete query string. Debug: If set then print some debug info.The default query is:
SELECT htmID_6,GSChtm FROM regions LIMIT 100
- messier_aitoff.pro: query the Messier table from the DIF DB and plot objects positions on an aitoff sky projection.
Note:- we assume coordinates are in degrees,
- you need to have the IDL Astronomy user’s library installed (https://idlastro.gsfc.nasa.gov/) and the custom routines loadct19 and aitoff_grid2 (available in the Soft/IDL directory).
The performed query is:
SELECT ra, decl, mag FROM Messier
- sid_mcshpdemo1.pro: use by default the ucac2_initial catalogue (assumed to be SID indexed with HEALPix order 8, column name healp8) in the mpe2018db DB to perform a SELECT query on a circular sky region and return the HEALPix pixel IDs (at order 8) of the objects.
The map will show the number of occurrences per pixel (we refer to it as the Nocc map – see the healp_MapFill function).
By default Mollweide and orthographic maps are produced with equatorial coordinate system.
Used routines: DBExec_Qry, healp_MapFill, healp_MapSave. It accepts the following input keywords:Host: The DB server host name (def. localhost). User: The user name to use for DB access (def. mpeusr). Password: The password to use for DB access (def. mpe2018pass). DBName: The database name to access (def. mpe2018db). TABName: The catalogue table name (def. ucac2_initial) Order: The map Order (resolution level). Nested: If set then assume NESTED scheme (used only if Order kwd passed). OutFile: If set then save in that file (def. into "mcsdb_map.fits"). Units: Units of output data (used for the plot). Gnomic: If set then plot Gnomic view (see gnomview help). Debug: If set then print some debug info. PNG: If set then save map into PNG files (see mollview help). Any extra keyword is passed to mollview / orthview / gnomview.And the default query is:
query1 = 'CALL SID.SelectCircleHEALP("myregion_circ", "*", "'+ dbname +'.'+ tabname +'", "healp8", 8, "RAmas/3.6e6", "DECmas/3.6e6", 0, 0, 1500, "")' query2 = 'SELECT HEALPlookup(0,8, ramas/3.6e6, decmas/3.6e6) as healpID_nest_8 FROM SID.myregion_circ'which is a circular region of 1500 arcmin radius, around RA, Dec (0,0). HEALPix map or order 8 is used (about 1 square deg pixels). Note that can save this map into an output FITS file using the OutFile keyword.
- mcshpdemo1.pro: this assumes DIF is installed! Use by default the ucac2_initial catalogue (assumed to be DIF indexed with HMT depth 6) in the mpe2018db DB to perform a SELECT query on a circular sky region and return the HEALPix pixel IDs (at order 8) of the objects.
The map will show the number of occurrences per pixel (we refer to it as the Nocc map – see the healp_MapFill function).
By default Mollweide and orthographic maps are produced with equatorial coordinate system.
Used routines: DBExec_Qry, healp_MapFill, healp_MapSave. It accepts the following input keywords:Host: The DB server host name (def. localhost). User: The user name to use for DB access. Password: The password to use for DB access. DBName: The database name to access. Query: The complete query string (see code for def.). Note that it has to produce a one column output! TABName: The reference table name (i.e. without the "_htm" or "_healp" extensions) used to get auxiliary info like the map Order/Nside. If not given then must give the Order keyword when the Query is an input. Order: The map Order (resolution level). Can give the TABName instead. Nested: If set then assume NESTED scheme (used only if Order kwd passed). OutFile: If set then save in that file (def. into "mcsdb_map.fits"). Units: Units of output data (used for the plot). Gnomic: If set then plot Gnomic view (see gnomview help). Debug: If set then print some debug info. PNG: If set then save map into PNG files (see mollview help). Any extra keyword is passed to mollview / orthview / gnomview.And the default query is:
SELECT HEALPlookup(0,8, ramas/3.6e6, decmas/3.6e6) as healpID_nest_8 FROM ucac2_initial_htm_6 WHERE DIF_Circle(0, 0, 1500)which is a circular region of 1500 arcmin radius, around RA, Dec (0,0). HEALPix map or order 8 is used (about 1 square deg pixels). Note that can save this map into an output FITS file using the OutFile keyword.
- mcshpdemo2.pro: this assumes DIF is installed! Like for mcshpdemo1 but use a default SELECT query that returns not only the number of occurrences of objects per pixel but also their magnitude.
The result will be two maps: the usual one with the number of objects per pixel, and the Sum map with the data of the second parameter of the query, in this case the object magnitude.
Note the usage of the /NORMpix keyword in the call to healp_MapFill. This asks the function to normalize the Sum map by the Nocc map. The result is a map with the average magnitude per pixel. This is the map that will be plot.
Edit the surce code if you want to chenge this.
The default query is:SELECT HEALPlookup(0,8, ramas/3.6e6, decmas/3.6e6) as hp8_ID, Amm/1e3 as Amag FROM ucac2_initial_htm_6 WHERE DIF_Circle(0, 0, 1500)Can try other queries with different regions, circular or rectangular, or with a different pixel size, or even a full sky one.
In the last case use the original table (ucac2_initial), not the view, which is needed by the DIF functions.
Remember to give the Order and eventually the TABName keywords. Also your query need always to return 2 columns, with the second one being normalized by the first one and then plotted. - mcshpdemo3.pro: very much like mcshpdemo1 but does not assume that table has a HEALPix column and plot the orthographic sphere in shade view. Edit the code at your wish.
The default query is:SELECT HEALPLookup(0,8, RAmas/3.6e6, DECmas/3.6e6) as kpx8 FROM ucac2_initial_htm_6 WHERE DIF_Rect(10, 20, 1200) - hprectsel.pro: this assumes DIF is installed! rectangular selection of objects on a given table. Default selection rectangle is
[10,30,240,120], which means a rectangle centered at RA,Dec (10,30) with sides of 240 arcmin along RA and 120 arcmin along Dec. The map is plotted in gnomic projection.
Input keywords:TABName: The reference table name (i.e. without the "_htm" or "_healp" extensions). Must be HTM indexed. Def. 'ucac2_initial'. Rect: a 4 elements array with the corner of the rectangle: [RAcen,DECcen,sideRA,sideDEC]. Def. [10,30,240,120] (deg,deg,',') OutFile: If set then save in that file (def. into "mcsdb_map.fits"). Any extra keyword is passed to "gnomview".
As you can see from the source code, communicating with the DB server is very simple. This is from dbexample1.pro:
@mcs_usrlib ; User contributed Functions and Pro PRO DBExample1 ; Open a database connection on localhost, a user name and password will be ; interactively requested, though they can be passed as paramaters ; (see DBConnect help) DBConnect, 'mpe2018db' ; Get the list of available databases: for a SELECT query, DBQuery returns a ; structure matching the returned data format unless the kwd EXEC_ONLY is set sql = 'SELECT * FROM INFORMATION_SCHEMA.SCHEMATA' rec = DBQuery(sql, NREC=nrec) PRINT, 'Available databases:' FOR i=0, nrec-1 DO $ PRINT, ' - ', rec[i].SCHEMA_NAME ; Create a test table (first drop it if exists) dummy = DBQuery('DROP TABLE IF EXISTS test_mcs') dummy = DBQuery('CREATE TABLE test_mcs (c CHAR(10), i INT, f FLOAT, UNIQUE(i))') ; Insert some records using simple INSERT queries dummy = DBQuery('INSERT INTO test_mcs VALUES("hello", 100, 2.3),("goodbye", 200, 4.6),("finish", 300, 9.2)') ; Retrieve table structure: DBQryStruct returns a structure rec = DBQryStruct('SELECT * FROM test_mcs LIMIT 1') ; Check that the returned structure matches the corresponding one on DB side PRINT, 'Table test_mcs structure: ' HELP, rec, /STRUCT ; Drop the test_mcs table dummy = DBQuery('DROP TABLE test_mcs') ; Close DB connection DBClose END
Examples of usage:
IDL>
dbexample1
simpleqry
dbexample_reg
dbexample_reg, user='mpeusr', pass='mpe2018pass', db='mpe2018db', $
query='SELECT * FROM mpe2018db.simpleBSC LIMIT 20'
messier_aitoff
mcshpdemo1, user='mpeusr', pass='mpe2018pass', db='mpe2018db'
mcshpdemo1, user='mpeusr', pass='mpe2018pass', db='mpe2018db', order=6, rot=[0,90], $
query='SELECT HEALPlookup(0,6, ramas/3.6e6, decmas/3.6e6) as hp6_ID FROM ucac2_initial_htm_6 WHERE DIF_Circle(82, -70, 600)'
mcshpdemo2, user='mpeusr', pass='mpe2018pass', db='mpe2018db'
mcshpdemo2, user='mpeusr', pass='mpe2018pass', db='mpe2018db', order=6, rot=[0,0], units='Amag', col=5, $
query='SELECT HEALPlookup(0,6, ramas/3.6e6, decmas/3.6e6) as hp6_ID, Amm/1e3 as Amag FROM ucac2_initial_htm_6 WHERE DIF_Rect(0, 0, 1200, 1800)'
mcshpdemo2, user='mpeusr', pass='mpe2018pass', db='mpe2018db', units='Amag', /gnomic
mcshpdemo3
hprectsel
hprectsel, rect=[13, -73, 500, 300]
Note how in the first query string we also added the DB name, which was not needed because it is identical to the DB we asked to connect to (mpe2018db).
In the second mcshpdemo1 we customized the query, pixel size and sky plot rotation.
We also specified that the output map order is 6. This is necessary for custom queries producing HEALPix IDs.
In mcshpdemo2 we also added a custom units label and changed the color palette.
Coordinates (82, -70) correspond to LMC, (13, -73) to SMC. Other possible coordinates: Galactic Center (266.4, -28.94), M31 (10.675, 41.268), M42 (83.75, -4.583).
Let’s now use the ASCC 2.5 catalogue to perform other queries and plot examples (can perform similar queries also for the UCAC2 catalogue; just consider possible memory limits):
IDL> ; Objects density per pixel for the ASCC 2.5 catalogue mcshpdemo1, user='mpeusr', pass='mpe2018pass', db='mpe2018db', tabname='ascc25', order=6, outfile='ascc_k6.fits', $ query='SELECT HEALPLookup(0,6, RAmas/3.6e6, DECmas/3.6e6) FROM ascc25' ; Replot in Galactic coords mollview, 'ascc_k6.fits', coord=['c','g'], grat=[30,30], title='ASCC order=6' ; Downgrade from Nside 64 (2^6) to 16 (2^4) and plot the boundaries of Healpix pixels loaddata_healpix, 'ascc_k6.fits', dummy, map64 ud_grade, map64, map16, nside_out=16, order_in='RING' mollview, map16, coord=['c','g'], title='ASCC order=4', hbound=16 ; Average B-V color per pixel for the ASCC 2.5 catalogue mcshpdemo2, user='mpeusr', pass='mpe2018pass', db='mpe2018db', order=8, outfile='ascc_b-v.fits', $ query='SELECT HEALPLookup(0,8, RAmas/3.6e6, DECmas/3.6e6), (Bmm-Vmm)/1e3 AS BmV FROM ascc25 WHERE Bmm>0 AND Vmm>0' ; Replot in Galactic coords mollview, 'ascc_b-v.fits', coord=['c','g'], grat=[30,30], title='ASCC B-V' ; Downgrade from Nside 256 (2^8) to 64 (2^6) and do change order (RING) loaddata_healpix, 'ascc_b-v.fits', dummy, map256 ud_grade, map256, map64, nside_out=64, order_in='RING', order_out='RING' mollview, map64, coord=['c','g'], title='ASCC B-V'
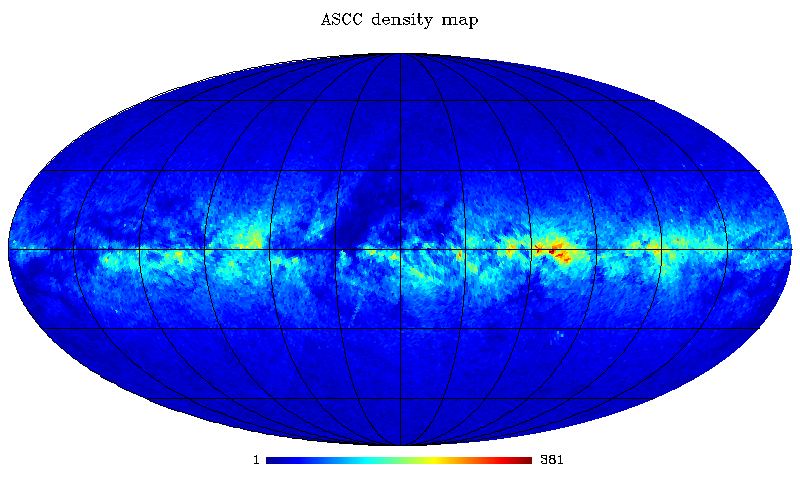
ASCC 2.5 star density – galactic Mollweide projection.
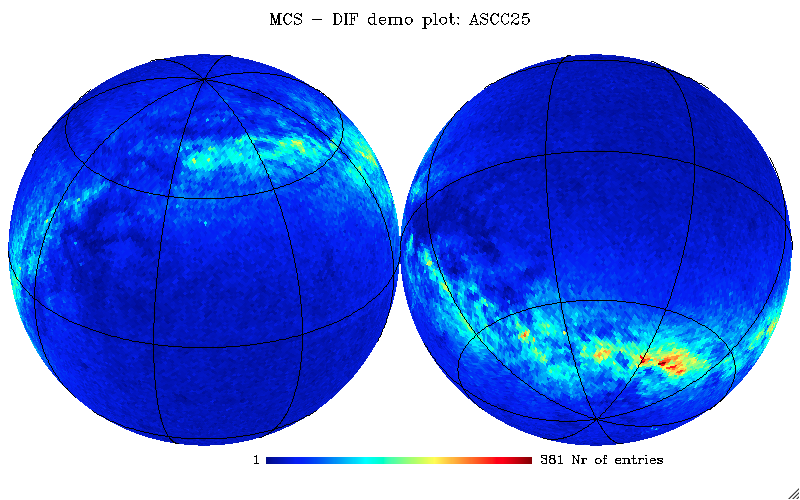
ASCC 2.5 star density – equatorial orthographic projection.
Note that the grey pixels can be seen in not full sky catalogues (like the UCAC-2). Those mark pixels with no value, i.e. NaNs. Can check bad pixels like this:
IDL> bad = where(finite(map256,/nan), nbad) IDL> print, nbad 98766
